Introduction
Limosilactobacillus fermentum is a lactic acid bacterium found in the human gastrointestinal tract and various fermented foods, and it has attracted interest for its health benefits as a probiotic [1,2]. This bacterium has the potential to improve gut health, enhance immune function, and provide antimicrobial effects [3]. However, the biological mechanisms supporting these effects and the specific genetic configurations of the strain are not yet fully understood [4]. Recent studies have focused on analyzing the genomic structure of L. fermentum to elucidate the relationship between its genetic diversity and physiological characteristics [5,6]. Whole-genome analysis, which includes the complete arrangement and structure of genes, provides fundamental information for understanding the microbial survival strategies, pathogenicity, adaptability, and biosynthetic capabilities [7]. Therefore, whole-genome data of L. fermentum play a crucial role in clearly understanding the health-promoting effects and interactions of the strain [8]. This paper aims to analyze the genetic characteristics of L. fermentum KL271 through its whole-genome and investigate how these characteristics contribute to its potential as a probiotic. This research intends to illuminate the physiological and genetic mechanisms of L. fermentum KL271 to facilitate the development and application of probiotics and contribute to the understanding of the complex interactions between the gut microbiota and human/animal health [9].
Meterials and Methods
To investigate the genomic characteristics of L. fermentum KL271, the bacterial strain was cultured under optimized conditions. Cultivation was performed in MRS broth at 37°C for 24 hours [10]. Genomic DNA was extracted using a commercially available kit (Exgene Cell SV Mini Kit, GeneAll, Korea) following the manufacturer’s protocol. For sequencing, the Illumina NovaSeq 6000 platform was utilized. Short-read libraries were prepared using the TruSeq Nano DNA Prep Kit, generating high-quality reads suitable for comprehensive analysis. Initial quality control of the raw sequence data was conducted using FastQC and FastP tools. Draft assembly was performed with Unicycler, followed by error correction using Pilon to enhance accuracy [11–13]. Genome annotation was achieved with Prokka for functional gene prediction. EggNOG mapper was employed to categorize gene functions and identify biological pathways [14]. To evaluate the assembly quality, QUAST metrics were utilized alongside BUSCO for benchmarking against universal single-copy orthologs. The resulting genome map was analyzed for features relevant to probiotic functionality, stress adaptation, and metabolic processes.
Results
The L. fermentum KL271 is 1,995,103 bp in length and comprises 1,928 coding sequences (CDS), with a high proportion of genes dedicated to metabolic processes and stress responses (Fig. 1). The genome also includes 4 rRNA genes, 55 tRNA genes, and 1 tmRNA gene, essential for protein synthesis and cellular function (Table 1).
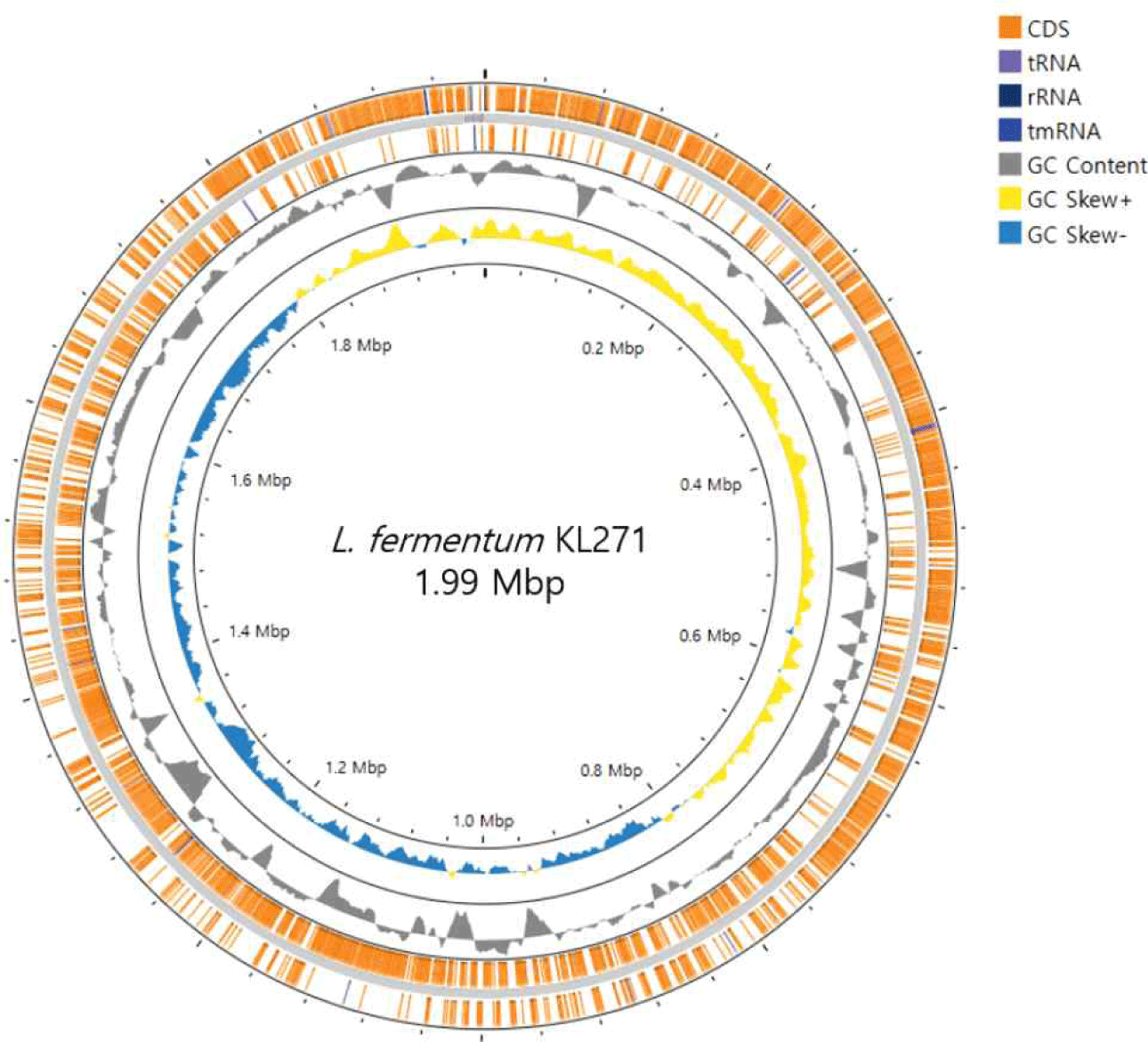
| Genomic features | L. fermentum KL271 (chromosome) |
|---|---|
| Genome size (bp) | 1,995,103 |
| GC content (%) | 51.36 |
| rRNA genes | 4 |
| tRNA genes | 55 |
| tmRNA | 1 |
| CDS | 1,928 |
Through genome analysis of L. fermentum KL271, various genes associated with probiotic traits have been identified. Notably, several genes related to the strain’s ability to adhere to the intestinal mucosa have been discovered. For example, genes such as Zinc import ATP-binding protein ZnuC and ABC transporter ATP-binding proteins endow KL271 with the capability to efficiently transport and absorb essential nutrients in the gut environment. These functions are crucial for the survival and proliferation of the microbiota in the intestine and play an important role in the interaction between KL271 and the host.
Additionally, KL271 possesses several genes that play a vital role in enhancing survival under stress conditions. The metabolic pathways involving Glutathione biosynthesis bifunctional proteins and Riboflavin biosynthesis protein RibBA contribute to reducing oxidative stress-related damage and maintaining cellular homeostasis [15]. These genes enable KL271 to effectively respond to stressful conditions in the gut environment, thereby enhancing its probiotic effects.
Genes involved in arginine and proline metabolism (15 genes) and glutathione metabolism (8 genes) were identified, which play significant roles in acid stress response and pH homeostasis (14 genes). These metabolic pathways help L. fermentum KL271 thrive in the gut environment.
4. CRISPR-Cas systems
The presence of CRISPR-Cas systems in L. fermentum KL271 was analyzed, revealing the diversity and potential adaptive immunity provided by these systems. Type I, II, III, VI, and IX CRISPR-Cas systems were identified, contributing to the strain’s ability to defend against phages and other genetic elements.






