Introduction
Fermented foods have long been recognized for their health benefits, with lactic acid bacteria (LAB) playing a crucial role in the fermentation process, contributing to the flavor, texture, and nutritional value of these foods [1]. Among LAB, Lactobacillus brevis is a significant species commonly found in various fermented products, such as kimchi, sauerkraut, and fermented beverages [2]. The ability of L. brevis to adapt to different environments and metabolize a wide range of substrates makes it an important microorganism for both food fermentation and potential probiotic applications [3]. Although L. brevis is not commonly used in fermented dairy products, recent studies have highlighted its potential as a starter culture in fermented milk applications due to its ability to produce gamma-aminobutyric acid (GABA), a bioactive compound with health-promoting properties, including stress reduction and blood pressure regulation [4]. Additionally, there are reports of its application in dairy fermentation processes as a complementary starter culture, contributing to both functional and sensory properties [5,6]. These studies suggest that L. brevis can play a dual role as a fermentative microorganism and a probiotic agent in dairy products. In this study, we analyzed the gene content and functional roles of L. brevis NS2301G3, a strain isolated from traditional fermented food, focusing on its contributions to carbohydrate metabolism, stress response, antimicrobial activity, and probiotic potential. The findings from this study highlight the versatility of this strain and its potential applications not only in traditional fermentation processes but also in the production of functional dairy products and other health-promoting foods.
Materials and Methods
Home-made kimchi samples were homogenized in sterile saline and plated on MRS (pH 5.0) agar [7]. Plates were then incubated anaerobically 37°C for 48–72 hours to allow for the growth of white colonies. Biochemical tests such as catalase test, Gram staining, and 16S rRNA gene analysis (Macrogen, Korea) were performed to initially identify the isolated strain. The strain identified as L. brevis, exhibiting high acid and bile tolerance, was designated as NS2301G3, and its whole genome was analyzed.
Two separate genomic DNA libraries were prepared according to the requirements of the Illumina and Oxford Nanopore systems. A combination of long-read Nanopore GridION and short-read Illumina Nextseq2000 platforms was used to generate the complete genome sequence of L. brevis NS2301G2. For Illumina sequencing, the extracted genomic DNA was fragmented by sonication using a Covaris M220 (Covaris, USA). The sheared DNA were then used to prepare a WGS library with an average insert size of 450 bp using a TruSeq Nano DNA Sample Prep kit (Illumina, USA). The library was sequenced on an Illumina Nextseq2000 platform (Illumina) using the 300 bp paired-end sequencing mode. For Nanopore sequencing, a MinION sequencing library was prepared using the Nanopore Ligation Sequencing Kit (SQK-LSK114; Oxford Nanopore, UK). The library was sequenced with an R10.4.1 GridION flow cell (Flongle) for a 24 h run using MinKNOW with the default settings (MinKNOW core 5.0.0, Guppy 6.0.6).
Illumina and Nanopore data were prepared for assembly, respectively with different options. Illumina Sequencing data were processed to remove low quality bases and adapter sequences with the optimized settings using Trimmomatic v0.39 (LEADING:10 TRAILING:10 SLIDINGWINDOW:4:20 MINLEN:200) [8]. Subsequently, additional phiX control were removed from pre-assembled data [9,10]. Trimmed sequences were aligned against phiX genome with bowtie2 v2.3.5.1 with the default options and filtered out by samtools v1.9 [11]. Nanopore sequencing data was basecalled with guppy basecaller v3.1.5. NanoFilt v2.8.0 was used to filter obtained reads with average Phred quality score lower than 7 and length lower than 1,000 [12,13].
Unicycler v0.4.8 was used to construct genome combined with Filtered NextSeq2000 and GridION data. After, genome was annotated using Prokka v1.14.6 and their coding sequences (CDS) were identified [14].
Results and Discussion
The basic genome statistics are provided in Table 1. The complete genome of L. brevis NS2301G3 consists of one circular chromosome (2,449,247 bp) with a GC content of 46.01%. According to the genomic results, L. brevis KL251 contains 2,412 CDSs, 67 tRNAs, and 15 rRNAs (Fig. 1).
| Genomic features | L. brevis NS2301G3 (chromosome) |
|---|---|
| Genome size (bp) | 2,449,247 |
| GC content (%) | 46.01 |
| rRNA genes | 15 |
| tRNA genes | 67 |
| CDS | 2,412 |
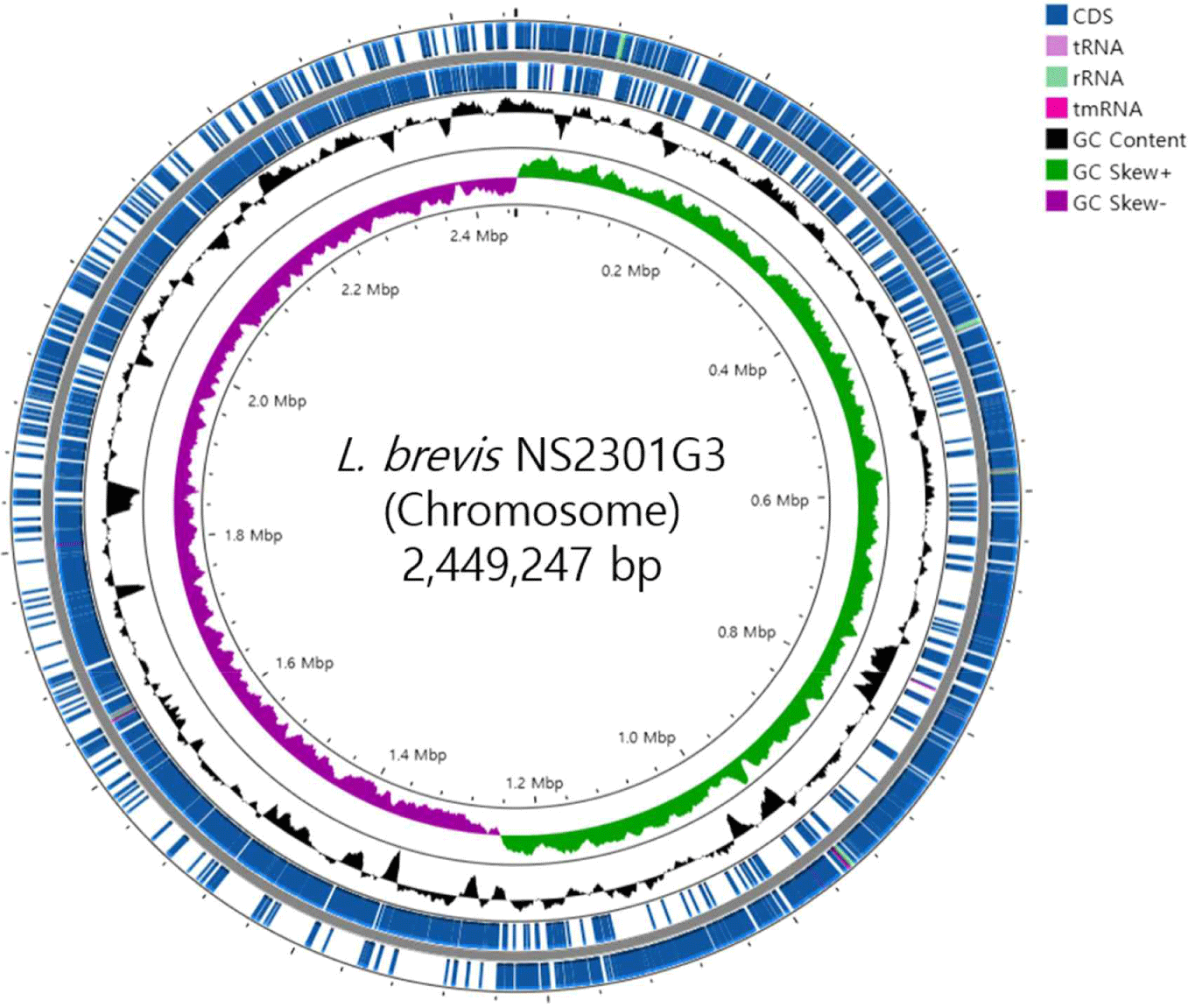
The complete genome of L. brevis NS2301G3 was analyzed to determine the gene content and its functional roles. Genome annotation was performed using the RAST server and Prokka annotation tools, providing an in-depth overview of the functional repertoire of this strain. The annotation revealed a total of 2,789 protein-CDSs, including genes involved in various metabolic pathways, stress response mechanisms, and cell structure maintenance. The annotated genome also identified several genes encoding carbohydrate-active enzymes (CAZymes), suggesting a strong potential for carbohydrate metabolism, which aligns with the species’ role in fermented foods [15]. Notably, genes responsible for the synthesis of lactic acid, such as lactate dehydrogenase (ldh), were prominently identified, emphasizing its importance in lactic acid fermentation [16]. Additionally, genes related to the catabolism of sugars such as glucose, fructose, and galactose were found, supporting the strain’s adaptability to diverse carbohydrate sources [17]. Key genes involved in carbohydrate metabolism include glucokinase (glk), phosphofructokinase (pfk), aldolase (ald ), beta-galactosidase (lacZ ), lactose permease (lacY ), and galactoside acetyltransferase (lacA) [18]. Functional annotation further revealed genes associated with stress tolerance, including those encoding heat shock proteins (DnaA, DnaN, DnaK, GroS, GroL) and oxidative stress response enzymes, which likely contribute to the strain's ability to survive and adapt to various environmental conditions, such as the acidic and anaerobic environment of fermented foods [19,20]. Additionally, genes involved in the biosynthesis of vitamins, such as riboflavin (ribBA, ribD) and folate (folT, folD, folC, folE, folK, folB), were detected, suggesting potential health-promoting properties [21,22]. In terms of antimicrobial properties, genes encoding CRISPR-associated proteins (casC, cas3), bacteriocins, and other antimicrobial peptides were found, indicating that L. brevis NS2301G3 may play a role in inhibiting pathogenic microorganisms in fermented food systems [23,24]. Genes associated with exopolysaccharide (EPS) production, such as pspA1, pspA2, pspB, were also identified, which are known to enhance the texture and viscosity of fermented products, thus contributing to the overall quality of the final product [25,26]. The genome also contains genes related to fatty acid metabolism regulation (fadR), arabinose metabolism repression (araR), and malolactic enzyme (mleS ), highlighting its versatility in metabolizing various substrates [27,28]. Genes involved in glutathione biosynthesis (gshAB), associated with antioxidant functions, were detected, further supporting the strain’s stress tolerance [29,30]. Additionally, genes related to the GABA pathway were identified, including glutamate/GABA antiporter (gadC ), glutamate decarboxylase (gadB), succinic semialdehyde dehydrogenase (gabD1), glutamate-tRNA ligase (gltX ), and proton/sodium-glutamate symporter (gltT ), suggesting the strain’s potential in GABA production, which is linked to health benefits such as stress reduction [31–33]. The presence of mobile genetic elements, including transposases and prophage sequences, was also noted, which may suggest genomic plasticity and the ability to acquire new traits through horizontal gene transfer [34]. This feature could provide an adaptive advantage in diverse ecological niches. Genes related to cell wall and mucosal adhesion, such as Eno, Eno2, LDH1, LDH2, LDH3, were also detected, which are likely important for the strain’s ability to interact with the host environment, contributing to its potential probiotic effects [35,36]. Overall, the gene content and functional annotation of L. brevis NS2301G3 reveal a versatile metabolic capacity, stress tolerance, and potential probiotic properties, supporting its application in fermented food production and its potential as a beneficial microbe for human health.






