Introduction
Whey proteins in milk have various functional properties and have been recognized for their biological value. These functional properties are particularly interesting to the food and pharmaceutical industries [1]. The major protein in whey is a crucial precursor of physiologically active peptides with various effects, such as α-lactalbumin and β-lactoglobulin cholesterol reduction. Moreover, whey protein contains immunoglobulins and serum albumin, which benefit the immune system when ingested [2].
Soybean is a widely acknowledged source of plant proteins, providing various health benefits. Besides proteins, soybean contains basic nutritive constituents, such as lipids, vitamins, minerals, free sugar, isoflavones, flavonoids, saponins, and peptides, which have therapeutic value [3].
In particular, studies have been conducted on the antioxidant activity and functionality of hydrolyzed whey- and soybean-derived peptides during lactic acid bacteria (LAB) fermentation. Furthermore, antioxidant activity varies depending on the amino acid sequence from the whey and soybean protein fermentation. Studies have revealed that whey- and soybean-derived proteins can serve as natural antioxidants and effectively suppress oxidation reactions in the food processing industry. Using these complex proteins can be challenging as they are highly resistant to hydrolysis and are hydrophobic, making it tasking to induce hydrolysis despite the precursor's high utility value. Peptide production through the fermentation of whey-derived proteins using LAB has gained attention in solving this challenge [4].
This study aimed to identify peptides with excellent anti-inflammatory and antioxidant properties in WPC and soybean protein fermentation. WPC and soybean protein fermentation result in post-degradation peptide conversion owing to proteolysis, causing improved functional properties. Enzymatic digestion of whey-derived proteins produces peptides from proteins [5,6]. However, enzymatic digestion is a high-cost hydrolysis method [7]. Therefore, hydrolysis of whey-derived proteins using LAB cultured on a large scale has been proposed as a lower-cost peptide production method [8]. This approach can be considered for producing peptides that display novel traits, which can be investigated for potential industrial applications [9].
Unlike other vegetable-based proteins, soybean has protein with digestible essential amino acid scores similar to WPC. Therefore, it is a high-quality protein among vegetable proteins. However, because WPC’s leucine content is higher than that in soybean protein, WPC is more effective in increasing muscle mass.
Materials and Methods
Whey protein concentrate (Table 1) was obtained from Meegle food Ingredients (MARQUEZ, USA). We added 5% (w/v) whey protein and glucose (Sigma-Aldrich, USA) at 5% (w/v) for LAB growth. It was adjusted to the optimum pH of the LAB with 1N NaOH (Sigma-Aldrich) and sterilization for 30 min at 80°C. The whey protein solution was heat-treated at 60°C for 60 min [10].
| Sample | Protein (%) | Moisture (%) | Fat (%) |
|---|---|---|---|
| WPC-80 | 83.36 | 4.50 | 6.90 |
Soybean (Table 2) is a new soybean type in Korea that has been improved for the effective adaptation of lactobacillus. After soaking soybeans at 15°C for 12 h, they were steamed for 7 min at 100±5°C, and we drained the water. After that, the purified water was wet-grinded by eight times the weight of the steamed soybeans. The okara was filtered through a 100-mesh filter to obtain 5±1 Brix soybean milk.
| Sample | Protein (%) | Carbohydrate (%) | Fat (%) |
|---|---|---|---|
| Soybean | 51.45 | 7.95 | 19.35 |
The ABTS radical cation scavenging activity of the ethanol extract was measured according to the method [11]. An ABTS stock solution was prepared by incubating a 7 mM ABTS solution dissolved in ethanol with 2.45 mM potassium persulfate for 16 h. ABTS solution was added and mixed with the sample at 9:1, and the absorbance was measured at 734 nm after 3 min [12]. Results are expressed in Trolox equivalent dose units (mM).
DPPH free radical scavenging activity was investigated by Huang et al. (2005) [13]. Samples were compounded with a 0.2 mM DPPH solution (Sigma-Aldrich) at 1:1 for the experiment and reacted for 30 min in a dark room. Absorbance was measured at 570 nm, and results were expressed as the ascorbic acid content (mM; Sigma-Aldrich).
The FRAP assay performed by Liu et al. (2013) [14] was used. The FRAP reagent contained 300 mM acetate buffer (pH 3.6):40 mM HCl solution:10 mM TPTZ:20 mM Iron(Ⅲ) chloride Hexahydrate at 100:9:1:10 (v/v). We mixed the FRAP solution with the sample at 19:1. Thereafter, the mixture was reacted for 30 min in a dark room. The absorbance was measured at 593 nm and expressed as the ascorbic acid content (mM; Sigma-Aldrich).
The hydroxyl radical scavenging activity of each sample was determined using the method described by Park et al. (2006) [15] with some modifications. Subsequently, the samples were mixed with 0.75 mM 1,10-phenanthroline (Sigma-Aldrich), phosphate buffer (pH 7.0), and 2.5 mM FeSO4 (Sigma-Aldrich) in a 1:1:1:1 ratio. Then, 20 mM hydrogen peroxide (Junsei Chemical, Japan) was finally mixed in the same ratio (v/v) as a reagent. After mixing, the mixture was reacted for 90 min in a dark room. Absorbance was measured at 593 nm, and results were expressed as the ascorbic acid content (mM).
Antibacterial activity was evaluated using the Paper disk method [16]. Pathogenic bacteria, including Escherichia coli (KCTC1682, Korea), Salmonella enterica (KCTC2054, Korea), Bacillus cereus (KCTC362A), and Staphylococcus aureus (KCTC3881, Korea), were obtained and dispensed into the Mueller Hinton agar (DifcoTM, USA). Then, 100 μL of the pathogenic bacteria culture (103–104 CFU/mL) was uniformly distributed by pouring 30 mL of pre-sterilized Mueller Hinton agar, which was allowed to solidify. A paper disk (Advantec, Japan) was impregnated with the compound and placed on the Mueller Hinton agar’s surface. The 100 μL protein sample complex was dispensed into each paper disk and incubated in a 37°C incubator. After incubation for 24 h, the diameter (mm) of the clear inhibition zone around the well was measured and compared.
Cell viability was determined using the MTT assay, as previously reported by Moon et al. (2019) and Peng et al. (2019) [17,18]. RAW 264.7 cells were seeded in 96-well cell culture plates for 24 h. After removing the medium, the RAW 264.7 cells were treated with various sample concentrations in a serum-free medium for 24 h. After that, the RAW264.7 cells were treated with MTT (5 mg/mL) for 2–3 h and re-incubated (37°C, 5% CO2). Then, the supernatants were removed. The dark blue formazan crystals formed in intact cells were extracted with DMSO, and absorbance was measured at 540 nm with a microplate reader (Bio-Tek Instruments, USA). Cell viability (%) was expressed in the treatment to control. The viability of cells was calculated considering controls as 100% viable.
RAW 264.7 cells were seeded in 96-well cell culture plates at 5×104 cells/well and incubated for 16–18 h. As explained previously, the NO quantity was estimated by measuring the nitrite, an oxidized product, in the cell culture supernatants. After removing the medium, RAW 264.7 cells were treated with various sample concentrations in the medium for 2–3 h. Thereafter, lipopolysaccharide (LPS) at a final 100 ng/mL concentration was treated and stimulated in the same medium for 20–22 h. Griess reagent was added to the supernatant at 1:1 (v/v). The absorbance was determined at 540 nm in a Microplate reader after 15 min at room temperature.
RAW 264.7 cells were seeded in 96-well plates at 5×104 cells/mL for 24 h, and pre-processed cell-free extracts were prepared. Then, the supernatant was removed, and the cytokine content was measured using the mouse interleukin (IL)-1α, IL-6, and tumor necrosis factor (TNF)-α ELISA kit (Komabiotech, Korea). Then 100 μL of the samples were added to each well coated with specific cytokine antibodies (IL-1α, IL-6, and TNF-α) and reacted at room temperature for 2 h. The supernatant was removed and washed five times with a washing buffer. Thereafter, a detection antibody was added to react with the antigen. Then, streptavidin-horseradish peroxidase conjugated with avidin was added and reacted at room temperature or 37°C for 30 min. After that, we aspirated and washed the plate four times. Then 100 μL of tetramethylbenzidine was added to each well as a substrate and incubated at room temperature for proper color development. A stop solution was added to each well, and absorbance was measured at 450 nm [10].
Amino acid profiling proceeded according to Waters amino acid analysis AccQ-Tag manual. Waters AccQ-Tag·Fluor reagent kit was used to derivate the sample and standard. Amino acid standard (Sigma-Aldrich) was used. The range of sample quantity was 0.02–0.08 μg (20–1,000 pmol). The other system conditions are presented in Table 3.
Peptide profiling of complex proteins was analyzed using HPLC (UltiMate 3000 system, Dionex, USA) and MS (Bruker Daltonics, 255748 Germany). Regarding the mobile phase, [H2O / FA = 100 / 0.2 (v/v)] was used as solvent A, and [Acetonitrile / FA = 100 / 0.2 (v/v)] was as solvent B. It was eluted for 46 min, and the gradient is presented in Table 4.
| Step | Total time (min) | Flow rate (μL/min) | A (%) | B (%) |
|---|---|---|---|---|
| 0 | 0 | 200.00 | 95.0 | 5.0 |
| 1 | 5 | 200.00 | 95.0 | 5.0 |
| 2 | 28 | 200.00 | 70.0 | 30.0 |
| 3 | 33 | 200.00 | 5.0 | 95.0 |
| 4 | 40 | 200.00 | 5.0 | 95.0 |
| 5 | 41 | 200.00 | 95.0 | 5.0 |
| 6 | 46 | 200.00 | 95.0 | 5.0 |
Results
Four antioxidant assays (ABTS, FRAP, DPPH, and hydroxyl radical scavenging) were conducted to verify antioxidant activity. The 8:2 sample had superior antioxidant activity to the control, confirmed through cross-validation (Fig. 1).

The sample that showed the highest inhibitory activity against all pathogens in the complex peptide sample in the conducted test was 8:2 (Table 5), showing similar or higher antibacterial activity than the positive control.
All samples showed high antibacterial activity against the pathogen S. aureus. The sample exhibited similar antibacterial activity on E. coli and S. enterica, indicated by a similar inhibitory region [19].
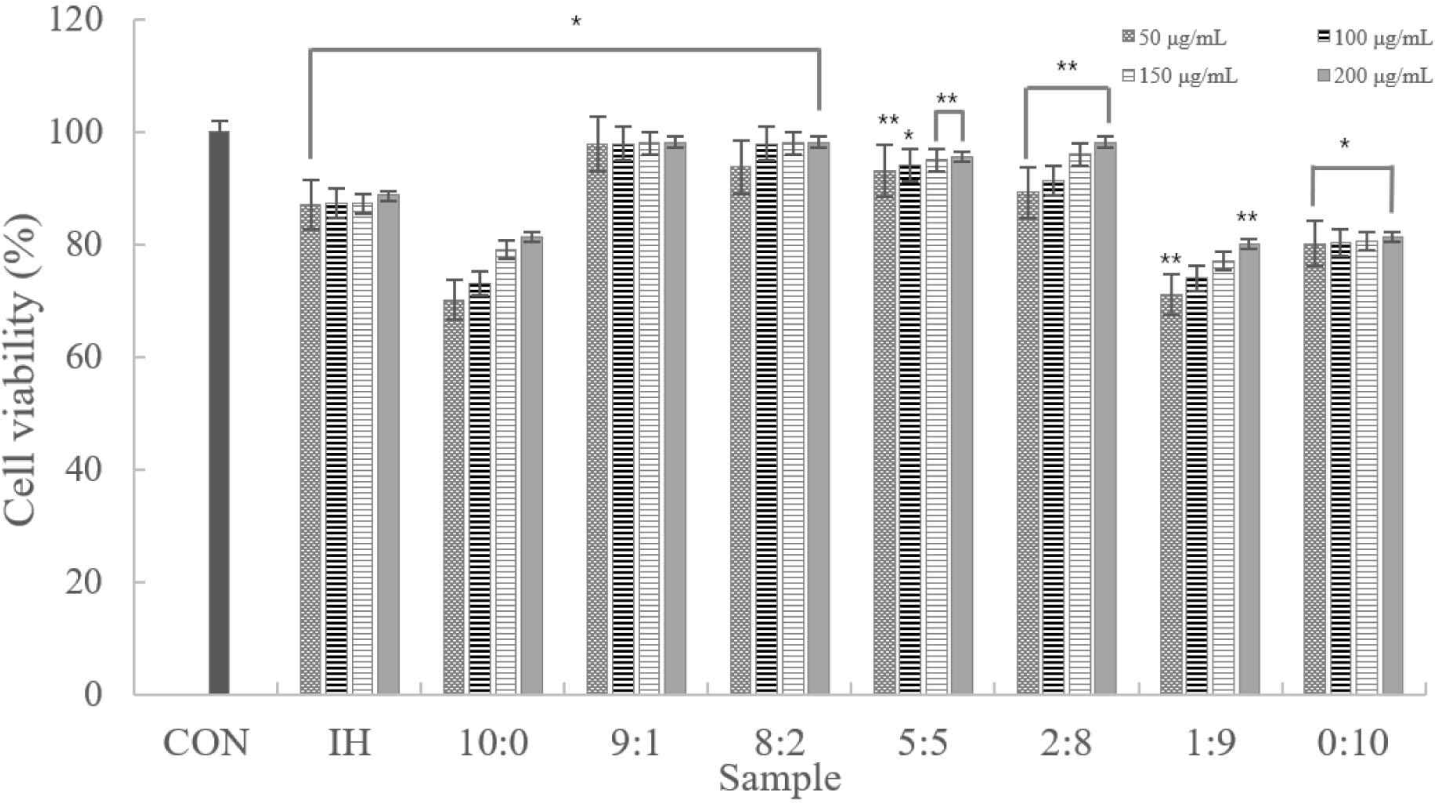
According to the experimental results, despite the increased concentration, there was no notable difference. Cytotoxicity was only observed in samples at 10:0, 1:9, and 0:10 ratios. In addition, samples other than those excluded had ≥80% viable cells. Therefore, we confirmed that the sample was not cytotoxic to the macrophages (Fig. 2).
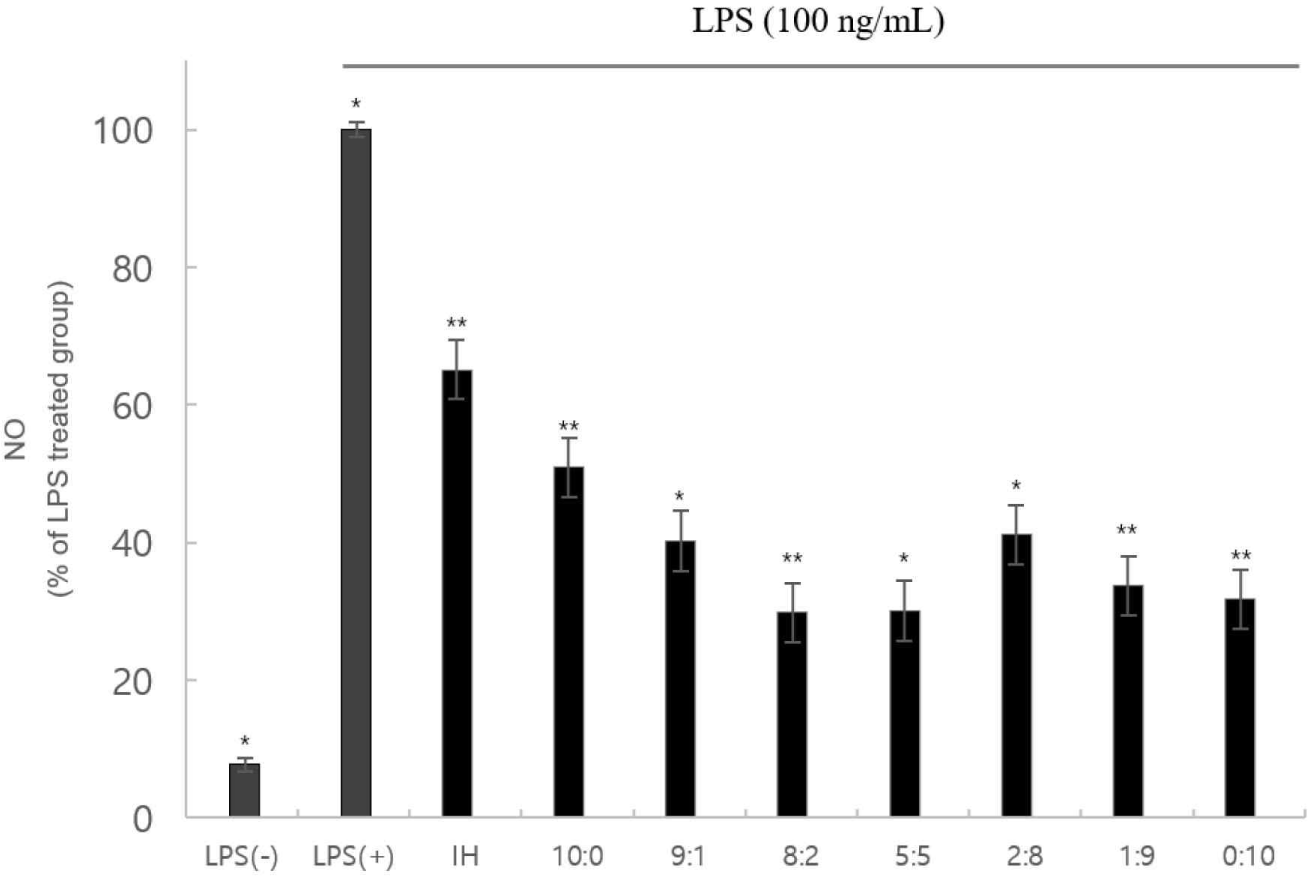
The NO assay revealed that cytotoxicity did not inhibit NO production (Fig. 3). These factors can functionally affect inflammation and cellular immunity positively.
IL-1α, IL-6, and TNF-α levels were measured, and the expression levels were plotted. Using ELISA, the cytokines (IL-1α, IL-6, and TNF-α) of the 8:2 sample were considerably lower than those of the LPS(+) group; the 8:2 sample had the lowest value (Fig. 4).
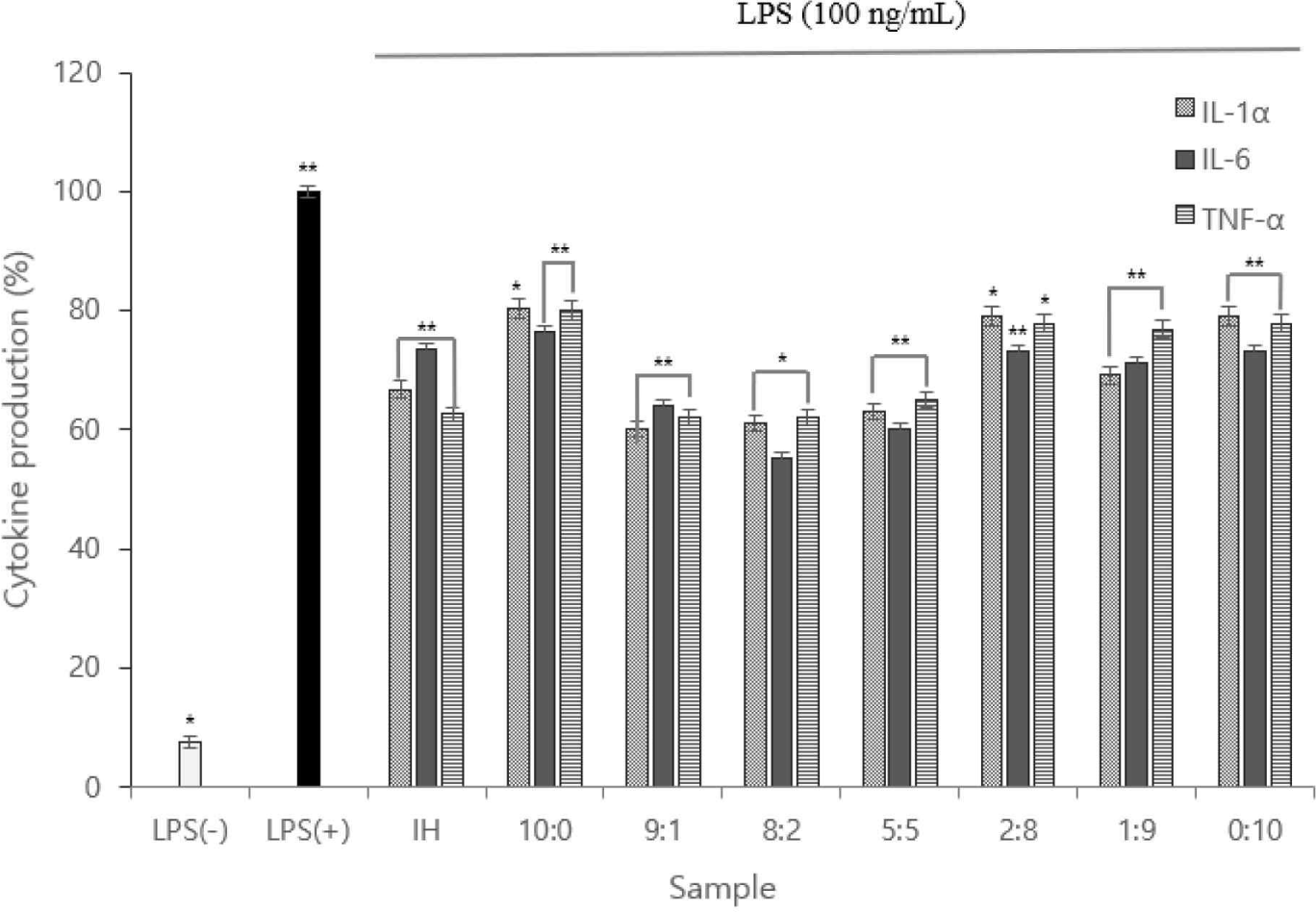
Therefore, we inferred the anti-inflammatory action of the complex peptides. Owing to the function of macrophages in each sample, the low TNF-α expression indicated that anti-inflammatory cytokine activation was inhibited.
Based on the cytokine measurement, the 8:2 sample had the best functional properties. This sample was a complex peptide fermented using Lactobacillus plantarum DK203 and Lactobacillus paracasei DK209. The AccQ-Tag system was used for the HPLC analysis of amino acids [10]. Seventeen amino acids were detected in the 8:2 sample (Fig. 5). Furthermore, the content ratios of phenylalanine and leucine were high. Glutamine and arginine—abundant in soybeans—also had large content ratios. The high ratios of phenylalanine and leucine were notable, as they are essential amino acids that require an influx of external nutrients.

WPC and soybean protein were subjected to LC-MS and LC-MS/MS for peptide separation and sequence identification, respectively. MS confirmed the presence of many other peptides. Several were fully sequenced and may require further purification. Additionally, six and two peptides are identified in Figs. 6 and 7, respectively. Tandem MS determined the molecular weight of the peptides. The characteristics of peptides identified using LC-MS/MS are listed in (Table 6).
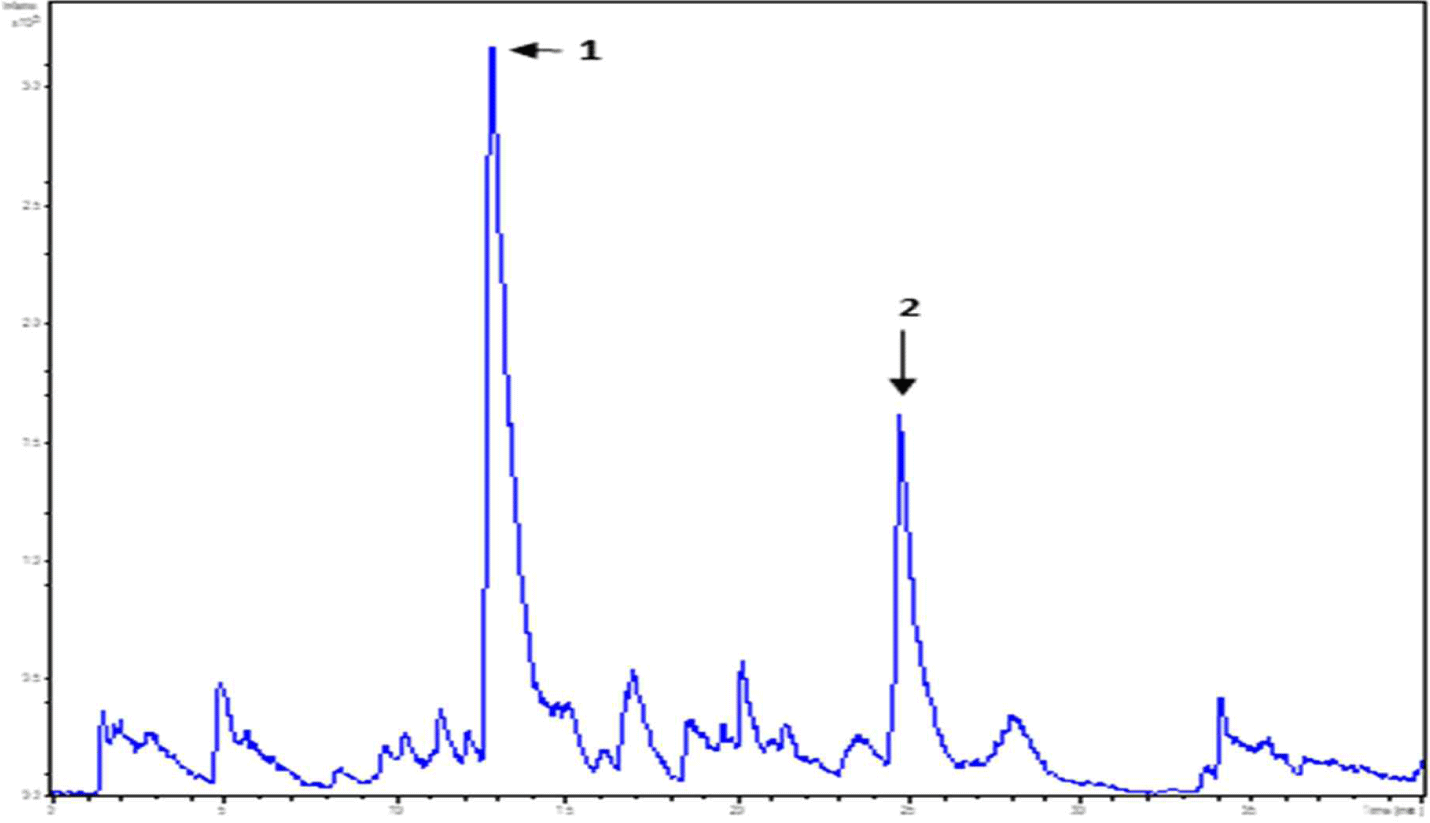
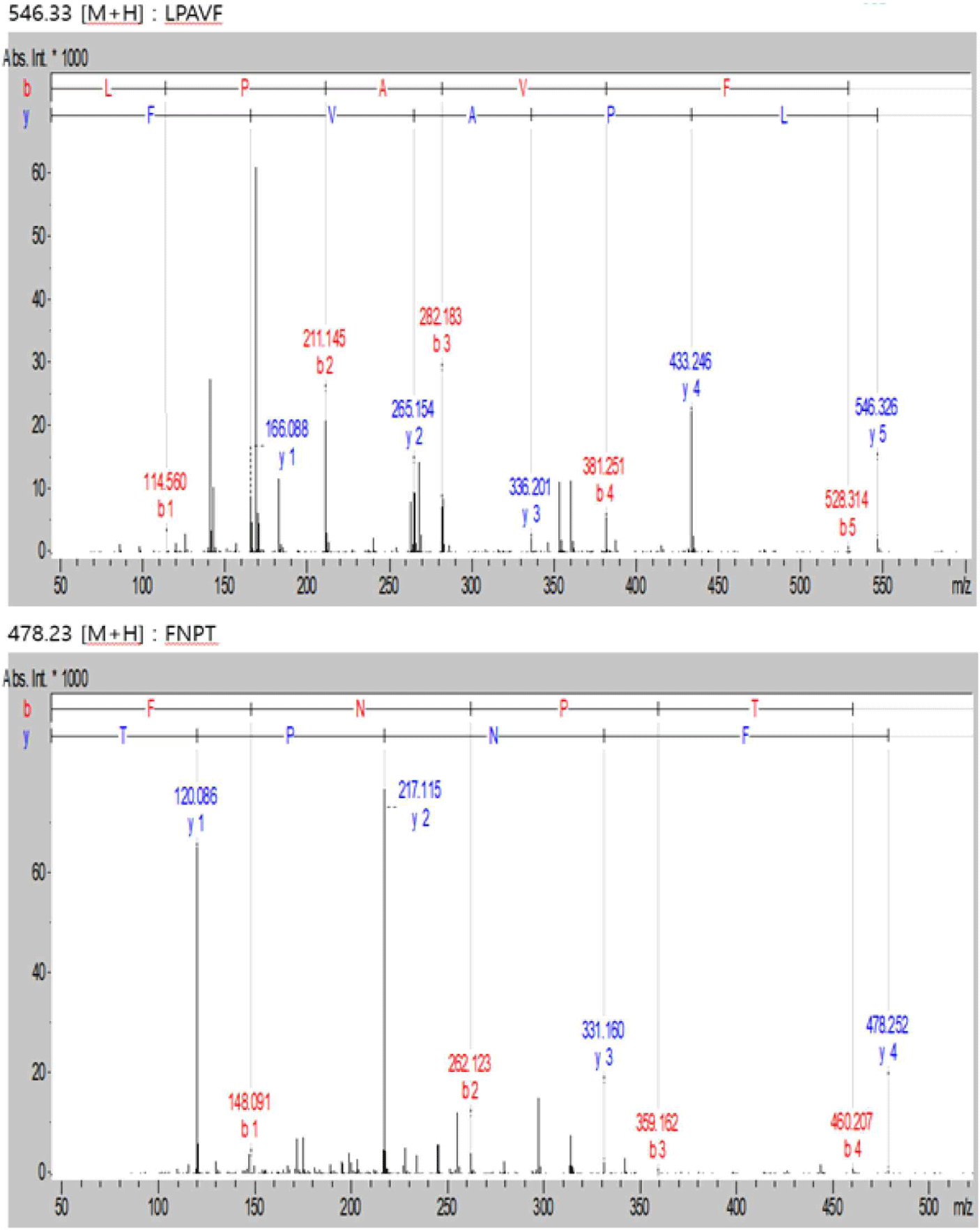
| Peak name | m/z | RT (min) | Sequencing |
|---|---|---|---|
| 1 | 478.23 | 12.8 | FNPT |
| 2 | 546.33 | 24.7 | LPAVF |
Discussion
Nine LAB strains from kimchi were isolated and identified. They were tested and stable up to pH 1.5. We observed that they strongly resist human digestive enzymes using artificial gastric juice and bile acid experiments.
Based on proteolytic activity and beta-galactosidase experiments, L. plantarum DK203, L. paracasei DK209, and L. paracasei DK215 strain were selected as candidate strains for whey protein hydrolysis. The two with the highest hydrolysis degree were selected among the candidate strains based on TNBS results. DK203 and DK209 were used as mixed strains as they performed the best regarding pH, titratable acidity, and viability after fermentation. The 8:2 sample had the highest antioxidant activity, as seen in the antioxidant activity assay.
In the antimicrobial experiment, 9:1 and 8:2 had higher antimicrobial activities than the control. All the samples demonstrated high antimicrobial activity against S. aureus.
The MTT assay indicated no significant difference in cell death, except for the 10:0, 1:9, and 0:10 samples. In addition, all samples other than the 10:0, 1:9, and 0:10 samples had ≥80 viable cells. Therefore, the sample was not cytotoxic to the macrophages. Regarding the NO assay, the lowest antioxidant activity occurred in the 8:2 and 5:5 samples. Cytokine (IL-1α, IL-6, and TNF-α) production in the 8:2 group was significantly lower than in the LPS(+) group. Therefore, anti-inflammatory activity was determined in the complex peptides. In addition, the low nTNF-α expression prevents the activation of pro-inflammatory cytokines produced by the expression of TNF-α.
The amino acid profile and sequence analysis of the 8:2 sample, which was the best, confirmed that the essential amino acid content was high. The amino acids Lys, Ile, and Cys, in varying contents, function as metabolic regulators for glucose metabolism in the body and affect body weight change [20].
Different cleavage sites may cause specific activity differences according to the LAB species, strain, and substrate differences. However, further studies are needed to determine the functional efficiency of specific LAB on whey and soy proteins. Thus, this study indicated that the complex fermented protein blends could serve as functional foods or dietary supplements, with commercialization planned.
Our tests hydrolyzing complex protein peptides with LAB confirmed that the functionality and content of free amino acids were increased. These properties show the applicability of LAB in the production of functional products utilizing dietary supplements, such as WPC and soybean protein. This indicates the efficiency of peptide mass production. Rather than producing complex protein peptides using expensive enzymes, the LAB fermentation method is a cost-effective alternative.
LAB can be cultured in large quantities with lower costs than enzyme production. However, a system that can control the physiological characteristics of the LAB is required. The biological characteristics of LAB may change depending on the culture conditions and environment, influencing the production of the target peptides. Controlling these conditions is crucial for efficiently producing complex peptides of uniform quality.






