Introduction
Milk is an essential source of nutrition for young mammals and contains several bioactive compounds essential for immune development and disease prevention [1,2]. Recent research has highlighted the presence of exosomes in the milk of several species, including cows [3–8]. These studies indicated that the cargo of bovine milk exosomes changes depending on the stage of lactation, with colostrum (the first milk produced after calving) particularly enriched in proteins that modulate immune responses and support neonatal development [9–11]. Encapsulated by a lipid bilayer approximately 100 nm in diameter, these exosomes are instrumental in cellular communication and immune responses and contain diverse contents such as lipids, proteins, mRNAs, and miRNA [8,12–16]. In addition, bovine colostrum exosomes (BCEs) show remarkable resistance to digestion and are being investigated as therapeutic agents for a variety of diseases (osteoporosis, inflammatory bowel disease, etc.) [10,15,17–19]. However, there remains a lack of comparative studies on the efficacy of exosomes derived from bovine colostrum and mature milk against both gram-negative and gram-positive bacterial pathogens.
C. elegans is a small soil nematode that is naturally exposed to numerous microorganisms [20]. In the context of immunity, C. elegans offers a valuable model for studying host-pathogen interactions. The nematode feeds on bacteria that pass through the pharynx into the gut, colonizing the gut lumen and forming the gut microbiota [21]. The nematodes interact closely with bacteria and have evolutionarily conserved defense mechanisms to evade pathogenic microbes [22,23]. Among the innate immune pathways in C. elegans, the PMK-1 pathway coordinates the defense against pathogens by regulating the expression of various antimicrobial effectors [20,24]. PMK-1 targets downstream genes that encode antimicrobial proteins, including antimicrobial peptides, C-type lectins (CLEC), and lysozymes [25]. Foodborne Pathogenic bacteria that are pathogenic to humans are also pathogenic to C. elegans, which has been reported as a useful model system in terms of pathogen-host interactions [20,26].
In this study, we used the C. elegans model to determine whether BCE induced immunomodulatory effects in pathogenic microbial infections. We found that BCE could trigger an innate immune response in C. elegans via the PMK-1 pathway. This protects worms from pathogenic bacterial infections and extends their lifespans. Our results provide an important basis for the further elucidation of the mechanisms underlying the protective effects of BCE against pathogenic bacterial infections in C. elegans.
Materials and Methods
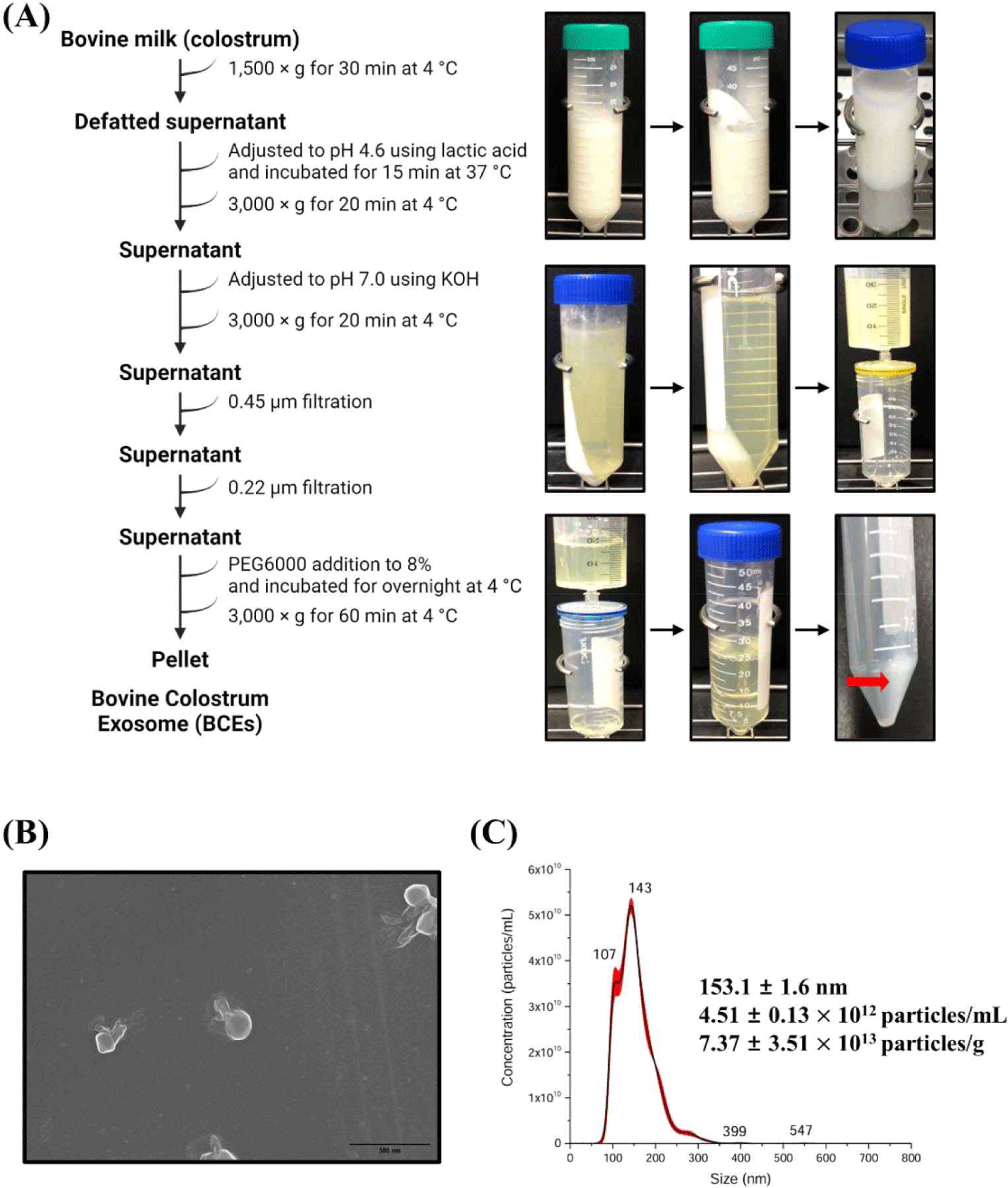
Bovine milk was collected from lactating milk samples (Korea). Milk samples were frozen immediately after milking and stored at –80°C until use. Exosomes were isolated from milk samples using established methods with modifications to previous studies [27,28]. Briefly, milk samples were centrifuged at 1,500×g at 4°C for 30 min to remove fat. The pH of the colostrum samples was adjusted to 4.6 using lactic acid and incubated for 15 min at 37°C to allow for maximum protein precipitation. The adjusted sample was centrifuged at 3,000×g for 20 min at 4°C to remove the casein. pH of casein- removed whey was adjusted to 7.0 using 5M KOH, then centrifuged at 3,000×g for 20 min at 4°C and the supernatant passed through a 0.45 and 0.22 μm filters in turn to remove residual cell debris. Polyethylene glycol (PEG) was added to the filtered whey to a final concentration of 8%. After addition of the PEG solution, the samples were thoroughly mixed and incubated overnight at 4°C. After overnight incubation, samples were centrifuged at 3,000×g for 1 h at 4°C. The conical tube was then decanted, drained for 5 min, and tapped occasionally to remove the excess PEG. PEG was removed, and the resulting pellet was resuspended in PBS (pH 7.4).
C. elegans N2 Bristol wild-type, CF512 fer-15(b26)II;fem-1(hc17)IV strains were used in this study. Worms were routinely maintained on NGM agar inoculated with OP50, a commonly used feed [29]. L1 worms were obtained by egg extraction from the sodium hypochlorite-sodium hydroxide solution of egg-bearing worms, then transferred to NGM plates seeded with HB101 and grown at 25 to obtain L4/young adult worms.
C. elegans longevity and killing assays were performed as previously described [29]. Briefly, bacteria were concentrated to 2×109 CFU/mL and plated in 100 μL aliquots on 35 mm-NGM agar plates. Milk exosomes (5 or 50 mg/mL) were mixed with concentrated bacteria and 100 μL aliquots were plated on 35 mm-NGM agar plates. The L4/juvenile adult and mutant N2 nematodes were individually transferred to each plate using a platinum wire and maintained at 25°C. Live worms were transferred to fresh plates daily during offspring production and viability was measured daily using a dissecting microscope (Olympus SZ40, Olympus, Japan). The worm was considered dead if it was not moved by gentle contact with a platinum wire pick. C. elegans killing analysis was performed as follows: L4/young adult worms were placed on plates exposed to milk exosomes or OP50 for 24 h at 25°C. Nematodes that had been pre-exposed to either milk exosomes (BCE) or OP50 were subsequently transferred to NGM plates inoculated with pathogenic bacteria. These nematodes were then monitored at 24 h intervals until mortality was complete, maintaining the temperature at 25°C.
Total RNA from C. elegans was extracted following the protocol provided with the TRIzol reagent (Invitrogen, USA) and subsequently purified using the RNeasy Mini Kit (Qiagen, Germany). After RNA extraction, cDNA synthesis was carried out using iScript cDNA synthesis kits (Bio-Rad, USA). For quantitative real-time PCR (qRT-PCR), the iTaq Universal SYBR Green Supermix (Bio-Rad) was used. The qRT-PCR were conducted on a StepOnePlus Real-Time PCR System (Applied Biosystems Applied). To determine the relative expression levels, the 2−ΔΔCT method was employed, with snb-1 serving as the reference gene for normalizing the gene expression data. RNA sequencing was provided by Macrogen (Korea).
We analyzed differentially expressed genes (DEG) by obtaining gene expression values through transcriptome sequencing of C. elegans, and performed functional classification and gene annotation for significant genes based on gene ontology (GO) and pathway information. The preprocessed trimmed reads were mapped to a known reference genome using HISAT2. After read mapping, transcript assembly was performed using StringTie. A DEG analysis was performed using these values. For the list of significant DEGs, we performed gene set enrichment analysis for biological processes (BP), molecular functions (MF), and cellular components (CC), which are functional classifications of GO, using gProfiler (https://biit.cs.ut.ee/gprofiler/orth).
Results and Discussion
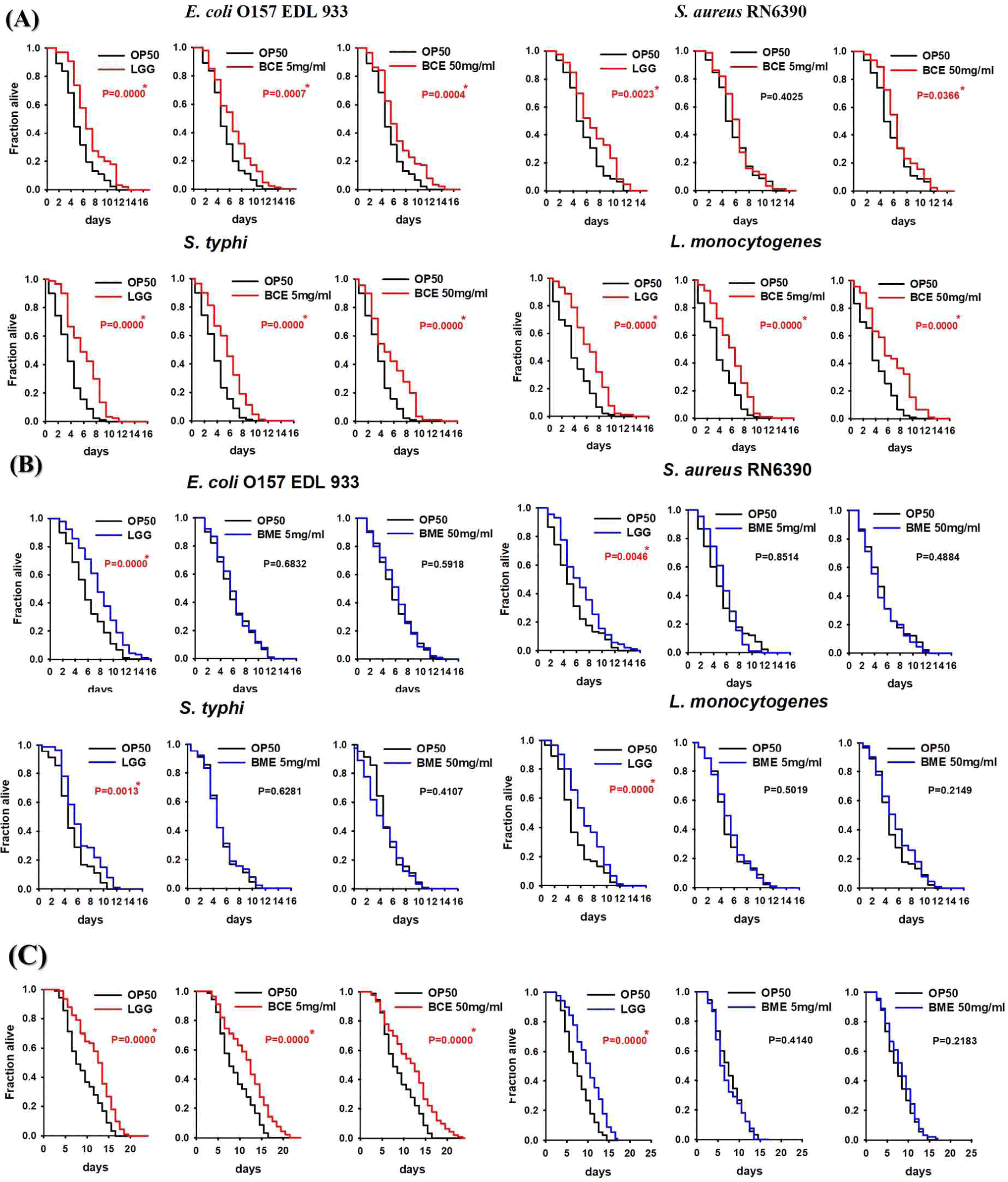
Cells release membrane vesicles called extracellular vesicles (EVs), which contain exosomes or microvesicles, into their extracellular environment [30,31]. EVs are found in various biofluids, indicating their functional importance [31,32]. Recent studies have shown that exosomes play an essential role in activating immune functions, and human milk exosomes have been shown to affect the immune system of infants [33]. Several studies have reported that the milk of mammals, including humans, is rich in proteins and miRNAs involved in immune responses [7,11,34]. Interestingly, early milk, such as colostrum, contains a higher concentration of exosomes than mature milk [35]. Exosomes were isolated from bovine colostrum using the PEG precipitation method, as depicted in Fig. 1A. The isolated exosomes were characterized using scanning electron microscopy (SEM) and nanoparticle tracking analysis (NTA). SEM confirmed the circular, membrane-enclosed shape of exosomes (Fig. 1B), while NTA measured the diameter of the exosomes at 153.1±1.6 nm. In addition, the particle concentration of the BCEs was approximately 4.51±0.13×1012 particles/mL, and the BCE yield was 7.37±3.51×1013 particles/g of BCE protein concentration (Fig. 1C). We used a C. elegans model to investigate the pathogen-host interactions of BCE against infection with pathogenic bacteria. First, we confirmed whether BCE exposure to four major foodborne pathogenic bacteria had an effect on lifespan compared to that of the control Escherichia coli OP50. The worms were exposed to BCE-plated plates for 24 h, followed by exposure to each of the four food-borne pathogenic bacteria. In a previous study, Lacticaseibacillus rhamnosus GG (LGG) was used as a positive control because it prolongs the lifespan of C. elegans and increases the survival rate by increasing resistance to pathogenic microbial infection [29]. E. coli OP50, an internationally established feed, was used as a negative control [36]. Worms exposed to 50 mg/mL BCE for 24 h showed improved survival against the four pathogenic bacterial infections compared with the control OP50 (Fig. 2A). Similarly, worms exposed to 5 mg/mL BCE for 24 h showed improved survival against three pathogenic bacterial infections. However, there was no significant difference in the survival rate compared to OP50 when infected with Staphylococcus aureus RN6390. Exosomes isolated from bovine colostrum milk have been shown to reduce virulence caused by infection with a foodborne pathogen in C. elegans. To confirm whether these results were the same for exosomes isolated from bovine mature milk, a killing analysis of C. elegans was performed using bovine mature milk exosomes (BME). Worms exposed to BME for 24 h showed no significant difference in survival rates for the four food-borne pathogen infections compared with those exposed to OP50 (Fig. 2B). Next, we evaluated whether BCE affects aging in C. elegans. The results confirmed that when BCE was used, the lifespan was significantly extended compared to that of OP50 (the control group). In contrast, when treated with BME, there was no significant difference compared to the control OP50 (Fig. 2C). These results suggest that BCE could extend the lifespan by enhancing the resistance of C. elegans to food-borne pathogenic infections.
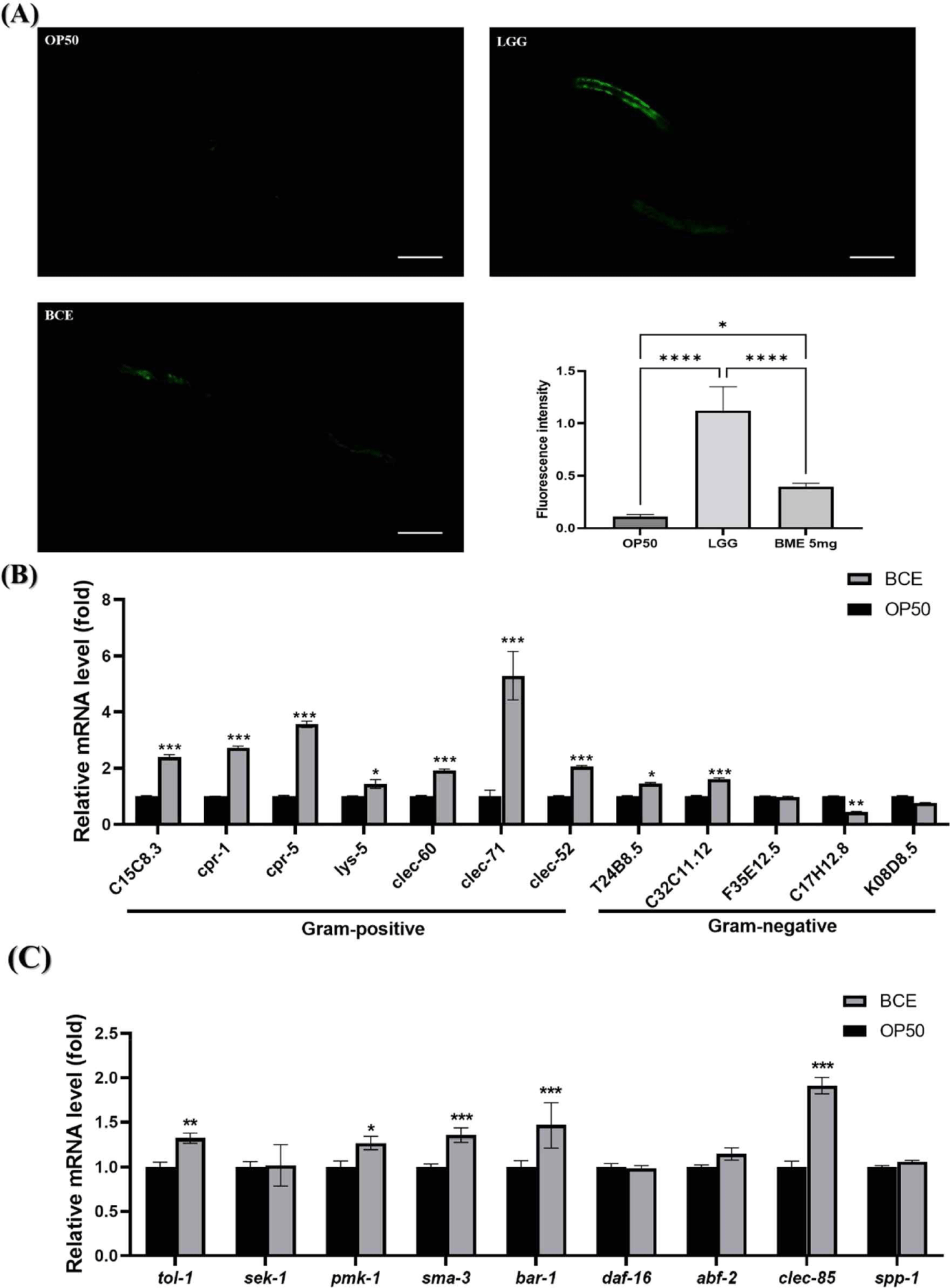
C. elegans has an innate immune system that responds to pathogenic bacterial infections by expressing defense genes [37]. We investigated the mechanisms by which BCE is associated with immunoregulatory and defense genes in C. elegans. PMK-1 is critical for the innate immune response and has been reported to be necessary for pathogenic bacterial infections [38]. We investigated whether BCE could regulate PMK-1 by using AY102 worms capable of confirming PMK-1::GFP expression. Our group previously identified that the LGG strain activates the PMK-1 pathway to enhance the susceptibility of C. elegans to pathogenic bacterial infections and used it as a positive control [29]. Although the fluorescence intensity of PMK-1 was weaker than that of LGG, BCE significantly increased the fluorescence intensity compared to that of the control OP50 (Fig. 3A). Based on our finding that prior exposure to BCE increases resistance to pathogenic bacterial infections, we investigated whether certain immune factors are modulated by exposure to BCE. We investigated 12 genes involved in resistance to pathogenic microbial infections identified in previous studies [29,37]. BCE treatment significantly increased the expression of 9 of the 12 genes (Fig. 3B). In particular, all genes related to gram-positive pathogens increased significantly, and five genes (C15C8.3, cpr-1, cpr-5, clec-71, and clec-52 ) increased more than two-fold. Next, we investigated the major genes (tol-1, sek-1, pmk-1, sma-3, bar-1, and daf-16 ) [39] involved in lifespan extension in C. elegans, and genes related to antibacterial peptides (abf-2, clec-85, and spp-1) [40]. The expression of pmk-1 was significantly increased by BCE treatment, as observed by fluorescence analysis. In addition to pmk-1, the tol-1, sma-3, and bar-1 genes were significantly increased, and among the antibacterial peptide-related genes, clec-85 was significantly increased. Immune response and longevity are closely linked in C. elegans (Fig. 3C). C. elegans possesses immune defense mechanisms, such as the p38 MAPK, transforming growth factor-beta (TGF-β), and beta-catenin signaling pathways [39]. In addition, several antimicrobial proteins produced by signaling pathways involved in defense against pathogen infections induce innate immune responses in worms [41]. Lysozyme (LYS) and CLEC families are antibacterial proteins in nematodes [42]. Several studies have reported that increasing the expression of these antimicrobial proteins prolongs the lifespan of nematodes during pathogenic infections [43,44]. These results suggest that BCE treatment prolongs the lifespan of patients with pathogenic infections by stimulating the innate immune response and increasing the production of antimicrobial proteins.
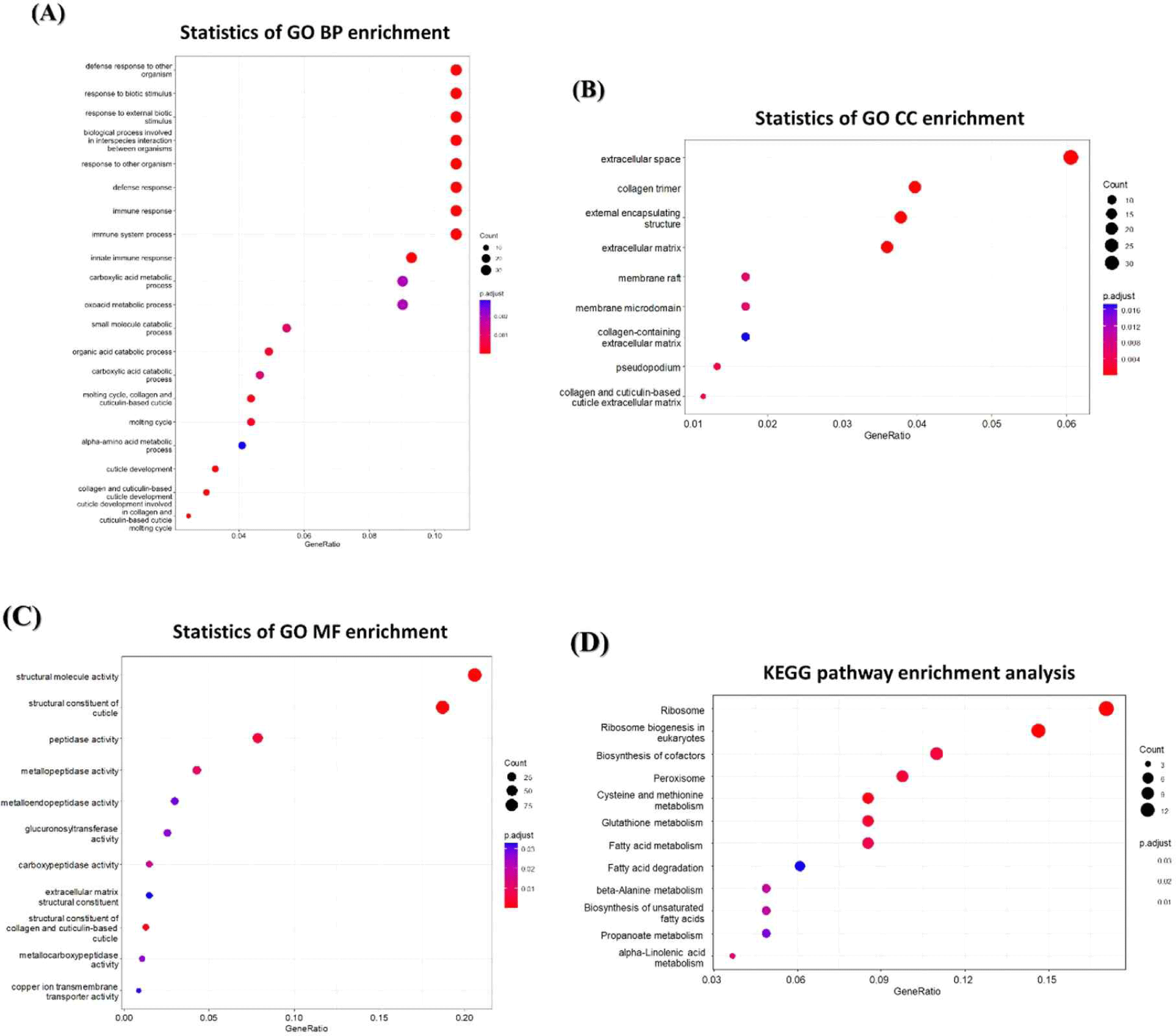
We used RNA-seq to analyze the changes in mRNA expression in C. elegans exposed to BCE. As a result, 1,677 genes were downregulated and 1,199 genes were upregulated, showing more than a 2.0-fold changes compared to the control groups. To investigate the genome-wide distribution in BCE-exposed C.elegans, a comprehensive GO enrichment analysis was performed. We identified 20 upregulated GO terms related to BP (Fig. 4A), nine associated with CC (Fig. 4B), and 11 related to MF (Fig. 4C). Consistent with the qRT-PCR results, BP-related GO enrichment analysis (Fig. 3) indicated that the significantly upregulated GO terms related to immunity in C. elegans exposed to BCE included defense responses to other organisms, defense responses, immune responses, immune system processes, and innate immune responses. In addition, we utilized DAVID (version 6.8) to identify the enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways for the mRNAs that changed following BCE exposure (Fig. 4D). Fatty acid metabolism may play a direct role in defense against multiple stresses, including heat, osmotic, and oxidative stress [45]. Anderson and Pukkila-Worley [46] reported that lipid metabolism is required for immune activation and pathogen defense in C. elegans. BCE have been reported to be significantly enriched in proteins involved in innate immune and inflammatory responses [11]. In addition, it was investigated that miRNAs related to immunity are also highly present in colostrum-derived exosomes [7]. When administered orally, bovine milk-derived EVs alter the gut microbiome and enhance gut immunity [47]. Therefore, exposure to BCE was found to have a protective effect on pathogenic bacteria and prolong lifespan by regulating innate immune stimulation and lipid metabolism in C. elegans.
In conclusion, we confirmed that BCE prolonged the lifespan of C. elegans by increasing its resistance to pathogenic bacterial infections. Furthermore, exposure to BCE induces a worm innate immune response by stimulating antimicrobial proteins (clec-85) and immune defense mechanisms (pmk-1, tol-1, sma-3, and bar-1). Taken together, our results confirmed the potential of BCE against aging and pathogenic microbial infections. However, further studies are needed to understand the detailed molecular mechanisms involved in enhancing innate immunity against the resistance to pathogenic microbial infections caused by BCE.
