Introduction
Levilactobacillus brevis is a Gram-positive, facultatively anaerobic lactic acid bacteria (LAB) belonging to the family Lactobacillaceae within the phylum Firmicutes. Formerly classified under the genus Lactobacillus, Levilactobacillus underwent taxonomic reclassification in 2020 as part of the Lactobacillus reorganization based on phylogenomic analysis. This reclassification aimed to resolve phylogenetic inconsistencies and enhance the precision of bacterial taxonomy [1].
L. brevis is known for the hetero-fermentative LAB, converting various carbohydrates into lactic acid, acetic acid, CO2 or ethanol during fermentation. L. brevis is catalase-negative and typically lack cytochrome oxidase activity, traits common among lactic acid bacteria. L. brevis is found in various ecological niches, including fermented foods, plant materials, soil, and the gastrointestinal tracts of humans and animals. L. brevis is a key lactic acid bacterium found in sauerkraut, pickles, and kimchi, contributing to its unique flavor and health-promoting properties [2,3]. L. brevis exhibits probiotic potential, capable of surviving and colonizing the gastrointestinal tract, modulating gut microbiota composition, and conferring health benefits to the host. Their antagonistic activity against pathogenic bacteria, production of antimicrobial substances (e.g., bacteriocins), and immunomodulatory effects further enhance their probiotic properties [4-6].
Genomic studies have provided insights into the genetic diversity, metabolic pathways, stress response mechanisms, and probiotic attributes of L. brevis. Advances in genome sequencing technologies have facilitated comparative genomic analyses, strain-specific trait elucidation, and targeted genetic engineering for improved probiotic and industrial strains.
In this study, we present the complete genome sequence of L. brevis KL251, obtained through whole sequencing techniques. Through detailed genomic analysis, we aim to elucidate the genetic basis underlying its fermentative capabilities, probiotic potential, and other relevant physiological traits.
Meterials and Methods
Home-made kimchi samples were homogenized in sterile saline and plated on MRS (pH 5.0) agar [7]. Plates were then incubated anaerobically 37°C for 48–72 hours to allow for the growth of white colonies. Biochemical tests such as catalase test, Gram staining, and 16S rRNA gene analysis (Theragen Bio, Korea) were performed to initially identify the isolated strain. The strain identified as L. brevis, exhibiting high acid and bile tolerance, was designated as KL251, and its whole genome was analyzed.
The genomic DNA of L. brevis KL251 was extracted using the Exgene Cell SV mini kit (GeneAll) accotding to the instructions of the manufacturer. The complete genome of L. brevis KL251 was sequenced at Teragen (Theragen Bio) using the Illumina Novaseq 6000 (Illumina, USA) platform with the TruSeq Nano DNA Sample Prep Kit 150PE (Illumina), which generates short reads. The quality of the assemblies for all genomes were assessed by using QUAST version 4.6.3 (GitHub, USA). Sequence assembly was performed using the following programs: The raw data underwent quality control using fastp (version 0.20.0), the draft genome was assembled with unicycler (version 0.5.0), and errors were corrected using pilon (version 1.24) was utilized [8-10]. The assembled sequences were then assessed using busco (version v5.5.0_cv1). The chromosome contig annotation were performed using rapid prokaryotic genome annotation Prokka (version 1.14.5-2). Using Eggnog-mapper (version 2.1.9) [11], we gathered functional annotations for genes or proteins.
Results
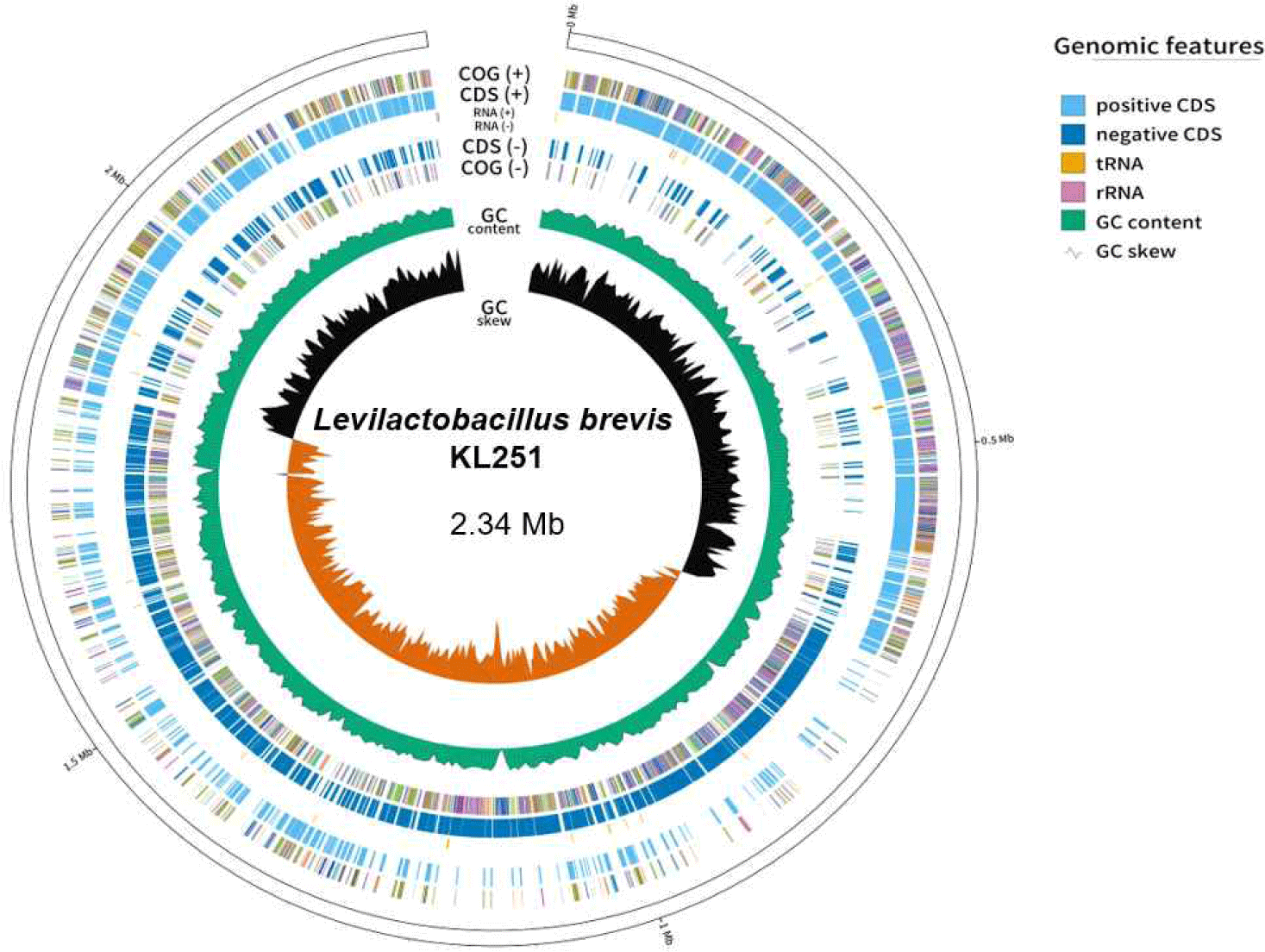
The basic genome statistics are provided in Table 1. The complete genome of L. brevis KL251 consists of one circular chromosome (2,345,062 bP) with a GC content of 45.78%. According to the genomic results, L. brevis KL251 contains 2,275 CDSs, 56 tRNAs, and 4 rRNAs (Fig. 1). Various associated genes have also been identified. It has genes (gadB_1, gadB_2, gadC_1, gadC_2, gadC_3) involved in GABA production, indicating its potential for GABA synthesis [12]. L. brevis KL251 has four CRISPR genes. One of them is associated with the type I-E CRISPR-Cas system, and the second one is associated with the type II CRISPR-Cas system, both of which are potentially active systems. The other two CRISPR gene loci serve as subunits. In addition, functional genes such as Isopentenyl-diphosphate Delta-isomerase, Glutathione amide reductase, and Redox- sensing transcriptional repressor (Rex) are involved in Isoprenoid biosynthesis, the generation of glutathione, and sensing the redox state based on the intracellular NAD+/NADH balance, respectively. They play crucial roles in regulating the expression of respiratory genes by detecting the oxidation-reduction (redox) state [13].
| Genomic features | L. brevis KL251 (chromosome) |
|---|---|
| Genome size (bp) | 2,345,062 |
| GC content (%) | 45.78 |
| N50 (bp) | 2,272,047 |
| rRNA genes | 4 |
| tRNA genes | 56 |
| tmRNA | 1 |
| CDS | 2,275 |
These data suggest that KL251, due to its stability and abundance of functional genes, is expected to have a substantial impact on probiotics. The genetic information enhances our understanding of KL251’s involvement in fermentation and its potential as a probiotic culture in food and health-related applications.